Supplementary material
Philippe H., Lopez P.
Trends Biochem Sci. 2001 Jul;26(7):414-6
![]() Data
Set
Data
Set
The GAPDH data set can be downloaded here in PHYLYP format, and the ADH data set can be downloaded here.
![]() Heterotachous
positions
Heterotachous
positions
Heterotachous positions are mapped on the human GAPDH (PDB entry 3GPD) with the following RasMol script. Here is the list of the hetrotachous positions numbered from the start of the human GAPDH:
28, 30, 31, 34, 36, 46, 47, 48, 52, 57, 59, 70, 71, 74, 78, 86, 90, 100, 103, 105, 120, 126, 128, 148, 159, 160, 161, 162, 163, 169, 171, 173, 180, 185, 190, 192, 193, 194, 195, 214, 222, 225, 226, 230, 231, 241, 245, 256, 257, 262, 274, 275, 279, 296, 300
![]() Phylogenetic
analyses:
Phylogenetic
analyses:
Two main phylogenetic analyses were performed for this study (ADH and GAPDH), both through a Neighbor-Joining reconstruction of a Kimura distance matrix. The corresponding trees are displayed below. Archaea are indicated in green, Eubacteria in brown and Eukaryotes in blue.
The following tree was reconstructed from 185 ADH sequences of 291 amino acid characters. Sequences from E. coli are indicated by a red arrow. The mixing of the colors in the tree clearly shows that horizontal gene transfers between domains are common for this gene. Moreover, the group of prokaryotes on page 1 only comprises cyanobacteria, alpha- and gamma-proteobacteria, and more generally shows a phylogeny that is not congruent with the usual bacterial phylogeny, which suggests that horizontal transfers also occur within a domain.

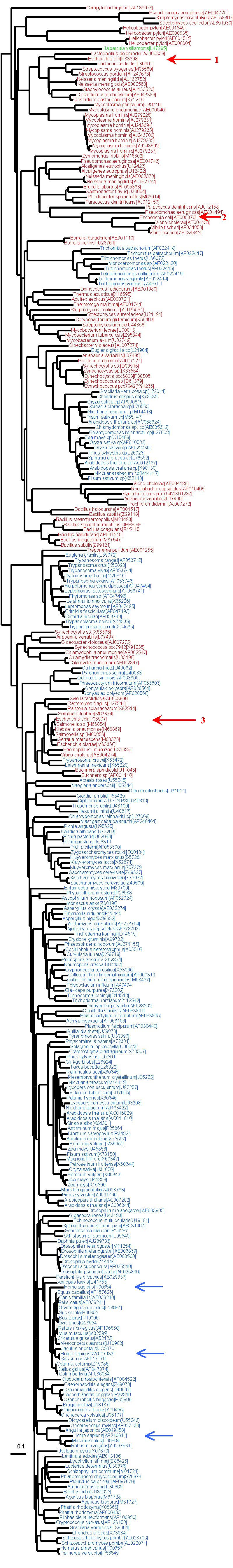
The following tree was reconstructed from 303 GAPDH sequences of 278 amino acid characters. Sequences from H. sapiens are indicated by a blue arrow, while E. coli sequences are indicated by a red arrow. The E. coli sequence number 3 was the one used by Kisters-Woike et al. and likely comes from an horizontal gene transfer. Sequence number 2 has likely changed its function (to D-eryhthrose apparently), and thus was not used in our manuscript. The “proper” bacterial-like sequence is indicated by red arrow number 1. The mixing of the colors shows that horizontal gene transfers are frequent between eukaryotes and bacteria for this gene. Moreover, the bacterial phylogeny that is recovered here is not congruent with the usual bacterial phylogeny, meaning that horizontal transfers also occur within domain, which makes the interpretation more difficult.