![]() This
table gives indices for all the trees used in the paper.
This
table gives indices for all the trees used in the paper.
Alpha Tubulin
| Data Set | Chars | MP steps | Cons. steps | Trees | C.I. | R.I. | Isym |
|
|
411
|
1346
|
1354
|
36
|
0.657
|
0.760
|
0.485
|
|
|
398
|
1206
|
1219
|
60
|
0.661
|
0.773
|
0.482
|
|
|
385
|
1092
|
1147
|
612
|
0.642
|
0.757
|
0.489
|
|
|
365
|
931
|
949
|
24
|
0.683
|
0.797
|
0.481
|
|
|
343
|
776
|
783
|
96
|
0.719
|
0.826
|
0.469
|
|
|
314
|
615
|
617
|
16
|
0.754
|
0.855
|
0.475
|
|
|
264
|
395
|
402
|
65
|
0.836
|
0.903
|
0.431
|
|
|
188
|
160
|
179
|
882
|
0.788
|
0.883
|
0.337
|
|
|
111
|
32
|
48
|
42
|
0.625
|
0.833
|
0.270
|
EF-1alpha
| Data Set | Chars | MP steps | Cons. steps | Trees | C.I. | R.I. | Isym |
|
|
388
|
1541
|
0.592
|
0.630
|
|||
|
|
326
|
833
|
843
|
10
|
0.637
|
0.686
|
0.485
|
|
|
308
|
691
|
693
|
3
|
0.659
|
0.711
|
0.557
|
|
|
281
|
514
|
529
|
40
|
0.711
|
0.729
|
0.486
|
|
|
250
|
349
|
364
|
297
|
0.769
|
0.774
|
0.459
|
|
|
220
|
230
|
231
|
2
|
0.866
|
0.868
|
0.422
|
|
|
168
|
92
|
97
|
31
|
0.907
|
0.924
|
0.318
|
|
|
109
|
15
|
15
|
3
|
1.000
|
1.000
|
0.229
|
Small subunit RNA (transversions only, except first row)
|
|
|
|
|
|
|
|
|
|
|
875
|
5043
|
5120
|
3432
|
0.29
|
0.628
|
0.302
|
|
|
875
|
2200
|
2208
|
1128
|
0.254
|
0.659
|
0.356
|
|
|
801
|
1369
|
1436
|
> 10000
|
0.329
|
0.702
|
0.300
|
|
|
771
|
1143
|
1245
|
8671
|
0.368
|
0.717
|
0.245
|
|
|
743
|
972
|
1021
|
7964
|
0.403
|
0.733
|
0.199
|
|
|
697
|
747
|
787
|
1062
|
0.463
|
0.772
|
0.206
|
|
|
630
|
502
|
524
|
2076
|
0.556
|
0.822
|
0.158
|
|
|
526
|
231
|
232
|
479
|
0.758
|
0.906
|
0.122
|
|
|
376
|
25
|
25
|
2
|
1.000
|
1.000
|
0.063
|
The common symmetry index from Heard et al. (Evolution 1992, 46:1818-1826) could not be used in our study because it has to be computed on perfectly dichotomic trees. We thus had to develop another index, in order to evaluate the symmetry of our many multifurcated trees.
We defined the order of emergence of a taxon as the number of nodes between that taxon and the root. For the purpose of our study, we defined the index of symmetry as the sum of the order of emergence of all taxa. This sum was then normalized (divided by n(n+1)/2 - 1, where n is the number of taxa) so that the maximal value is 1 (perfectly asymmetric tree).
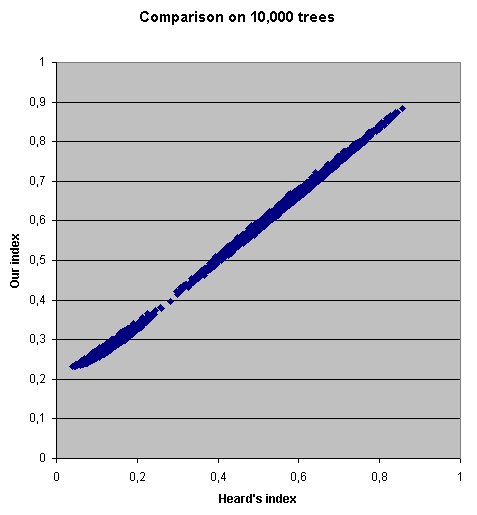
We performed a comparison between the two indices, for 10,000 random dichotomic trees of 50 taxa. The result showed a strong positive correlation between the two indices, meaning that they provide a similar order relationship for dichotomic trees. As can be observed, our index does not reach 0 for dichotomic tree, hence the negligible plateau in the lower left corner.
Though less intuitive than Heard's, our index allowed
evaluate the symmetry of multifurcating tree. We nevertheless had to postulate
that a radiation of 2n taxa (index 0) is more symmetric than
the perfectly symmetric 2n taxa tree (index 2(n-1)/((2n+1)-1)
). Another disadvantage of our index is that one cannot compare the symmetry
of trees with different numbers of taxa (as our index does not reach 0
for dichotomic trees).
Back to summary